Statistical Rethinking 2nd edition Homework 3 in INLA
library(tidyverse)
library(rethinking)
library(dagitty)
library(INLA)All three problems below are based on the same data. The data in data(foxes) are 116 foxes from 30 different urban groups in England. These foxes are like street gangs. Group size varies from 2 to 8 individuals. Each group maintains its own (almost exclusive) urban territory. Some territories are larger than others. The area variable encodes this information. Some territories also have more avgfood than others. We want to model the weight of each fox. For the problems below, assume this DAG:
hw3dag <- dagitty("dag{
avgfood <- area
weight <- avgfood
weight <- groupsize
groupsize <- avgfood
}")
plot(hw3dag)## Plot coordinates for graph not supplied! Generating coordinates, see ?coordinates for how to set your own.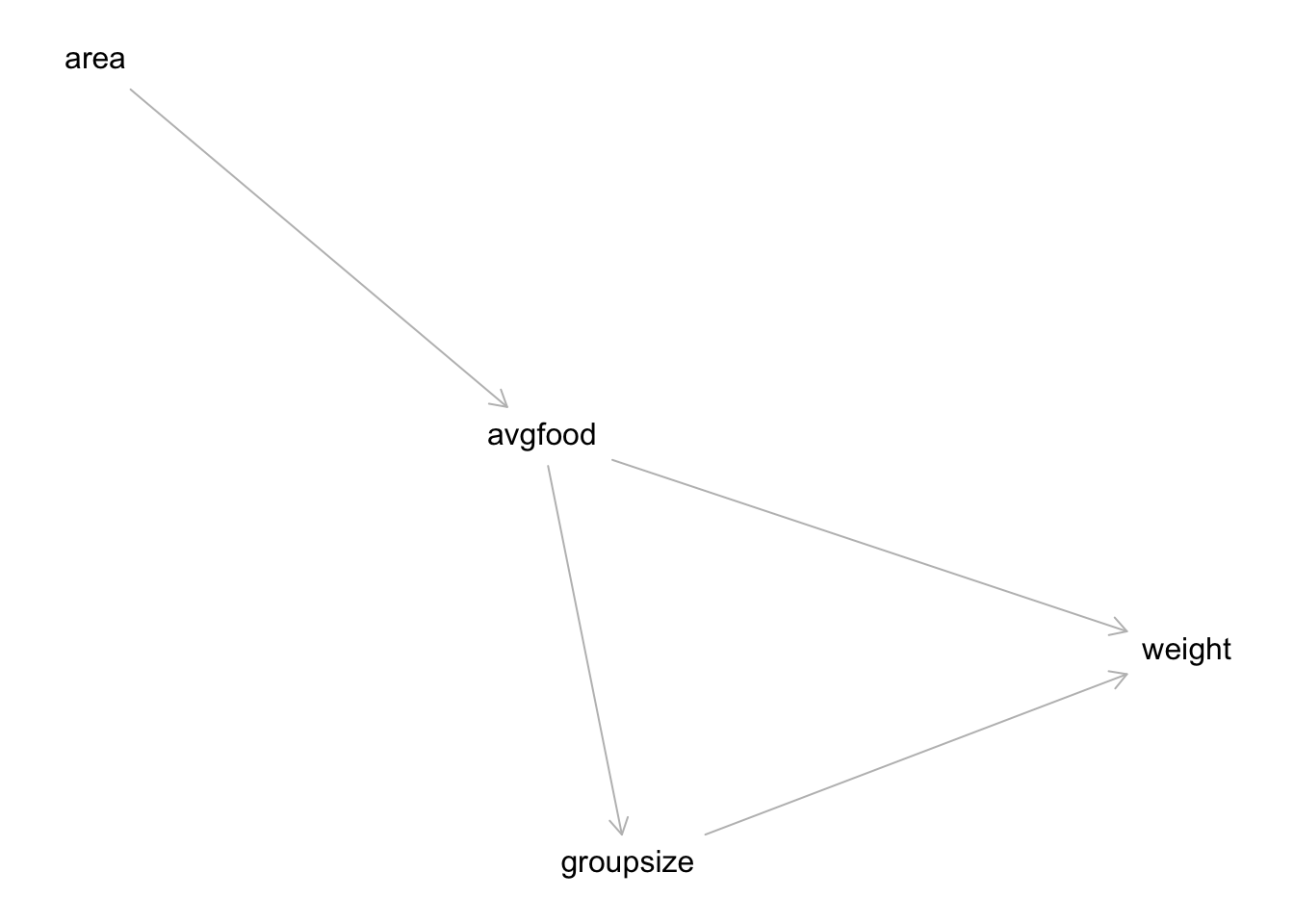
1. data(foxes) infer the total causal influence of area on weight
Use a model to infer the total causal influence of area on weight. Would increasing the area available to each fox make it heavier (healthier)? You might want to standardize the variables. Regardless, use prior predictive simulation to show that your model’s prior predictions stay within the possible outcome range.
1. rethinking
Because there are no back-door paths from area to weight,we only need to include area. No other variables are needed. Territory size seems to have no total causal influence on weight, at least not in this sample.
data(foxes)
f <- foxes
f$A <- scale( f$area )
f$F <- scale( f$avgfood )
f$G <- scale( f$groupsize )
f$W <- scale( f$weight)
m1 <- quap(
alist(
W ~ dnorm( mu , sigma ) ,
mu <- a + bA*A ,
a ~ dnorm( 0 , 0.2 ) ,
bA ~ dnorm( 0 , 0.5 ) ,
sigma ~ dexp( 1 )
) , data=f )
precis(m1)## mean sd 5.5% 94.5%
## a -2.483642e-05 0.08360740 -0.1336456 0.1335959
## bA 1.886254e-02 0.09089420 -0.1264040 0.1641290
## sigma 9.912479e-01 0.06466352 0.8879031 1.09459271. INLA
default priors
By default, the intercept has a Gaussian prior with mean and precision equal to zero. Coefficients of the fixed effects also have a Gaussian prior by default with zero mean and precision equal to
0.001. The prior on the precision of the error term is, by default, a Gamma distribution with parameters 1 and 0.00005 (shape and rate, respectively) ( so this is different from the statistical rethinking book’s prior on the variance which is exp(1))
#using default priors
m1.inla <- inla(W~A, data= f)
summary(m1.inla)##
## Call:
## "inla(formula = W ~ A, data = f)"
## Time used:
## Pre = 2.01, Running = 0.133, Post = 0.146, Total = 2.29
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) 0.000 0.093 -0.183 0.000 0.183 0.000 0
## A 0.019 0.094 -0.165 0.019 0.203 0.019 0
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for the Gaussian observations 1.01 0.132 0.767 1.00
## 0.975quant mode
## Precision for the Gaussian observations 1.28 0.992
##
## Expected number of effective parameters(stdev): 2.00(0.00)
## Number of equivalent replicates : 57.98
##
## Marginal log-Likelihood: -182.37The precision is the inverse of the variance. If we want to set the sd to 0.5, we have to set the precision to 1/(0.5^2)
m1.inla.prior <- inla(W~A, data= f, control.fixed = list(
mean= 0,
prec= 1/(0.5^2), # sd = 0.5 --> precision =1/variance --> 1/(sd^2)
mean.intercept= 0,
prec.intercept= 1/(0.2^2)
))
summary(m1.inla.prior)##
## Call:
## c("inla(formula = W ~ A, data = f, control.fixed = list(mean = 0, ", "
## prec = 1/(0.5^2), mean.intercept = 0, prec.intercept = 1/(0.2^2)))" )
## Time used:
## Pre = 1.17, Running = 0.133, Post = 0.136, Total = 1.44
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) 0.000 0.084 -0.166 0.000 0.166 0.000 0
## A 0.019 0.092 -0.162 0.019 0.199 0.019 0
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for the Gaussian observations 1.01 0.132 0.769 1.00
## 0.975quant mode
## Precision for the Gaussian observations 1.29 0.993
##
## Expected number of effective parameters(stdev): 1.79(0.025)
## Number of equivalent replicates : 64.84
##
## Marginal log-Likelihood: -177.652. data(foxes) infer the causal impact of adding food to a territory
Now infer the causal impact of adding food to a territory. Would this make foxes heavier? Which covariates do you need to adjust for to estimate the total causal influence of food?
2.rethinking
To infer the causal influence of avg food on weight,we need to close any back-door paths. There are no back-door paths in the DAG. So again, just use a model with a single predictor. If you include groupsize, to block the indirect path, then you won’t get the total causal influence of food. You’ll just get the direct influence. But I asked for the effect of adding food, and that would mean through all forward paths.
Again nothing. Adding food does not change weight. This shouldn’t surprise you, if the DAG is correct, because area is upstream of avgfood.
# food on weight
m2 <- quap(
alist(
W ~ dnorm( mu , sigma ) ,
mu <- a + bF*F,
a ~ dnorm( 0 , 0.2 ) ,
bF ~ dnorm( 0 , 0.5 ) ,
sigma ~ dexp( 1 )
) , data=f )
precis(m2)## mean sd 5.5% 94.5%
## a 1.686958e-07 0.08360013 -0.1336090 0.1336093
## bF -2.421092e-02 0.09088496 -0.1694626 0.1210408
## sigma 9.911433e-01 0.06465848 0.8878066 1.09448012.INLA
INLA default priors
#using default priors
m2.inla <- inla(W~F, data= f)
summary(m2.inla)##
## Call:
## "inla(formula = W ~ F, data = f)"
## Time used:
## Pre = 1.19, Running = 0.132, Post = 0.139, Total = 1.46
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) 0.000 0.093 -0.183 0.000 0.183 0.000 0
## F -0.025 0.094 -0.209 -0.025 0.159 -0.025 0
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for the Gaussian observations 1.01 0.132 0.767 1.00
## 0.975quant mode
## Precision for the Gaussian observations 1.28 0.992
##
## Expected number of effective parameters(stdev): 2.00(0.00)
## Number of equivalent replicates : 57.98
##
## Marginal log-Likelihood: -182.36INLA custom priors
#using custom priors
m2.prec.prior <-
m2.inla.prior <- inla(W~F, data= f, control.fixed = list(
mean= 0,
prec= 1/(0.5^2),
mean.intercept= 0,
prec.intercept= 1/(0.2^2)
))
summary(m2.inla.prior)##
## Call:
## c("inla(formula = W ~ F, data = f, control.fixed = list(mean = 0, ", "
## prec = 1/(0.5^2), mean.intercept = 0, prec.intercept = 1/(0.2^2)))" )
## Time used:
## Pre = 1.22, Running = 0.131, Post = 0.149, Total = 1.5
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) 0.000 0.084 -0.166 0.000 0.166 0.000 0
## F -0.024 0.092 -0.205 -0.024 0.156 -0.024 0
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for the Gaussian observations 1.01 0.132 0.769 1.00
## 0.975quant mode
## Precision for the Gaussian observations 1.29 0.994
##
## Expected number of effective parameters(stdev): 1.79(0.025)
## Number of equivalent replicates : 64.84
##
## Marginal log-Likelihood: -177.643. data(foxes) infer the causal impact of groupsize
Now infer the causal impact of groupsize. Which covariates do you need to adjust for? Looking at the posterior distribution of the resulting model, what do you think explains these data? That is, can you explain the estimates for all three problems? How do they go together?
3. rethinking
The variable groupsize does have a back-door path, passing through avgfood. So to infer the causal influence of groupsize, we need to close that path. This implies a model with both groupsize and avgfood as predictors.
It looks like group size is negatively associated with weight, controlling for food. Similarly, food is positively associated with weight, controlling for group size. So the causal influence of group size is to reduce weight—less food for each fox. And the direct causal influence of food is positive, of course. But the total causal influence of food is still nothing, since it causes larger groups. This is a masking effect, like in the milk energy example. But the causal explanation here is that more foxes move into a territory until the food available to each is no better than the food in a neighboring territory. Every territory ends up equally good/bad on average. This is known in be- havioral ecology as an ideal free distribution.
# groupsize on weight
#need to include avgfood
m3 <- quap(
alist(
W ~ dnorm( mu , sigma ) ,
mu <- a + bF*F + bG*G,
a ~ dnorm( 0 , 0.2 ) ,
bF ~ dnorm( 0 , 0.5 ) ,
bG ~ dnorm( 0 , 0.5 ) ,
sigma ~ dexp( 1 )
) , data=f )
precis(m3)## mean sd 5.5% 94.5%
## a -9.170948e-05 0.08013639 -0.1281651 0.1279817
## bF 4.772661e-01 0.17911961 0.1909984 0.7635339
## bG -5.735298e-01 0.17913821 -0.8598273 -0.2872324
## sigma 9.420204e-01 0.06174870 0.8433340 1.04070673. INLA
INLA default priors
#using default priors
m3.inla <- inla(W~ F + G,family = c("gaussian"), data= f)
summary(m3.inla)##
## Call:
## "inla(formula = W ~ F + G, family = c(\"gaussian\"), data = f)"
## Time used:
## Pre = 1.18, Running = 0.133, Post = 0.157, Total = 1.47
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) 0.000 0.089 -0.174 0.000 0.174 0.000 0
## F 0.641 0.206 0.236 0.641 1.045 0.641 0
## G -0.739 0.206 -1.144 -0.739 -0.335 -0.739 0
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for the Gaussian observations 1.11 0.147 0.845 1.11
## 0.975quant mode
## Precision for the Gaussian observations 1.42 1.09
##
## Expected number of effective parameters(stdev): 3.00(0.00)
## Number of equivalent replicates : 38.66
##
## Marginal log-Likelihood: -181.14INLA custom priors
Prior mean for all fixed effects except the intercept. Alternatively, a named list with specific means where name=default applies to unmatched names. For example control.fixed=list(mean=list(a=1, b=2, default=0)) assign ‘mean=1’ to fixed effect ‘a’ , ‘mean=2’ to effect ‘b’ and ‘mean=0’ to all others. (default 0.0)
#using custom priors
m3.inla.prior <- inla(W~F+G, data= f, control.fixed = list(
mean= list(F=0, G=0),
prec= list(F=1/(0.5^2), G=1/(0.5^2)),
mean.intercept= 0,
prec.intercept= 1/(0.2^2)
))
summary(m3.inla.prior)##
## Call:
## c("inla(formula = W ~ F + G, data = f, control.fixed = list(mean =
## list(F = 0, ", " G = 0), prec = list(F = 1/(0.5^2), G = 1/(0.5^2)),
## mean.intercept = 0, ", " prec.intercept = 1/(0.2^2)))")
## Time used:
## Pre = 1.25, Running = 0.138, Post = 0.148, Total = 1.54
## Fixed effects:
## mean sd 0.025quant 0.5quant 0.975quant mode kld
## (Intercept) 0.000 0.081 -0.159 0.000 0.159 0.000 0
## F 0.474 0.181 0.116 0.475 0.827 0.477 0
## G -0.570 0.181 -0.923 -0.571 -0.213 -0.573 0
##
## Model hyperparameters:
## mean sd 0.025quant 0.5quant
## Precision for the Gaussian observations 1.11 0.147 0.843 1.11
## 0.975quant mode
## Precision for the Gaussian observations 1.42 1.09
##
## Expected number of effective parameters(stdev): 2.58(0.047)
## Number of equivalent replicates : 45.03
##
## Marginal log-Likelihood: -173.84